Writing a CROWN Configuration
The most important part of CROWN is the configuration file. In this file, all producers, cuts, shifts and output variables are defined. The configuration is contained in a configuration class. In the following, a simple example and the provided functions are given. A configuration has to include
Producers to be run
Parameters to be used by the producers
Output variables to be used
Additionally, it can include
Modifiers: Modifiers can be used to change parameters based on era or sample.
Rules: A modification rule can be used to add/remove producers based on era or sample.
Systematics: Systematics can be used to vary the parameters of the producers and add the varied output to the output file as well.
One advantage of CROWN is that several final states can be produced at the same time. First, the global scope is run, which includes all operations that are independent of the analysis’s final state and are identical for all final states. Then, the processing is split into several scopes, which are run in parallel. Each scope results in a separate output file in the end. All parameters, outputs, producers, modifiers and systematics are always defined, depending on the scope. The global scope can be treated as a normal scope, but it is always run first, and all other scopes inherit the parameters, producers, modifiers and systematics of the global scope.
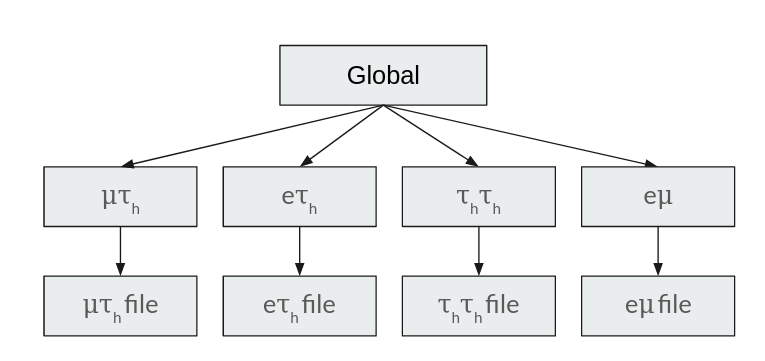
Basics
The configuration file should be contained in the config folder. During the build process, the user can specify the configuration file to be used by setting
-DANALYSIS=analysisname
where the analysisname is the name of the configuration file. Files in different folders can be specified using the Python import syntax e.g. -DANALYSIS=config.analysis.signal. The basic required structure of the configuration file is as follows:
def build_config(
era,
sample,
scopes,
shifts,
available_sample_types,
available_eras,
available_scopes,
):
configuration = Configuration(
era,
sample,
scopes,
shifts,
available_sample_types,
available_eras,
available_scopes,
)
#########################
# setup the configuration
#########################
...
#########################
# Finalize and validate the configuration
#########################
configuration.optimize()
configuration.validate()
configuration.report()
return configuration.dump_dict()
The configuration class provides several functions to set up a configuration conveniently. These functions will be explained in the following.
Parameters
Parameters are added using the add_config_parameters() function. A basic example usage of the function is
# add muon selection cuts
configuration.add_config_parameters(
["mm", "mt'],
{
"muon_iso_cut": 0.3,
"min_muon_pt": 23.0,
"max_muon_eta": 2.4,
"muon_id": "Muon_mediumId",
},
)
This adds the parameters muon_iso_cut, min_muon_pt, max_muon_eta and muon_id to the parameter list of the producers mm and mt. These parameters can be used by any producer in the mm and mt scope and will be set to the given values. So e.g. in the GoodMuonPtCut producer
GoodMuonPtCut = Producer(
name="GoodMuonPtCut",
call="physicsobject::CutPt({df}, {input}, {output}, {min_muon_pt})",
input=[nanoAOD.Muon_pt],
output=[],
scopes=["em", "mt", "mm"],
)
the parameter min_muon_pt will be set to 23.0.
Configuration Modifiers
Modifiers can be used, to conveniently change parameters of the configuration based on the era and the sample. Two main methods are available
SampleModifier- Modifies the parameters of producers based on the sample.EraModifier- Modifies the parameters of producers based on the era.
Both have a similar interface. The basic structure of how to include a modifier is
configuration.add_config_parameters(
["mm", "mt'],
{
"applyRecoilCorrections":
SampleModifier({"wj": True}, default=False),
},
)
In this example, the parameter applyRecoilCorrections is added to the parameter list of the producers mm and mt. Due to the configuration of the modifier, the parameter will be set to True for the samples of the type wj. For all other samples, the default value False is set.
Internally, the resolving of modifiers is done using the resolve_modifiers() function, which is called automatically by the add_config_parameters() function.
Output Quantities
All output quantities will be included in the output ntuple ROOT file. The output quantities are defined using the add_outputs() function. The basic structure is
configuration.add_outputs(
["mm", "mt"],
[
nanoAOD.event,
q.pt_1,
q.pt_2,
q.m_vis,
],
)
This example adds the output quantities event, pt_1, pt_2 and m_vis to the output of the producers mm and mt. There are two types of quantities available:
Quantity- A quantity produced by a CROWN producerNanoAODQuantity- A quantity from the input nanoAOD file
Both types can be used as output quantities. A Quantity has to be defined in the code_generation.quantities.output file and a NanoAODQuantity is defined in the code_generation.quantities.nanoAOD file.
Set of Producers
The set of producers to be run can be defined using the add_producers() function. The basic structure is
configuration.add_producers(
"mm",
[
muons.GoodMuons,
muons.VetoMuons,
pairselection.MMPairSelection,
pairselection.GoodMMPairFilter,
]
)
In this example, the producers GoodMuons, VetoMuons, MMPairSelection and GoodMMPairFilter are added to the list of producers mm. The producers added here can be both a Producer or a ProducerGroup. Also, the order of the producers is not important. Using the optimize() function, the producers’ ordering will be optimized, such that filters are always run first, and that producers that depend on other producers are run after the producers that they depend on.
The collection of available producers can be found in the code_generation.producers folder. An explanation of how producers are set up and linked to their corresponding C++ function is given in Defining a New Python Producer.
Systematic Variations
Systematic Variations are an important part of physics analysis. CROWN provides a powerful way to set up systematic variations. The basic structure of including a systematic variation to the configuration is the add_shift() function. The basic structure is
configuration.add_shift(
SystematicShift(
name="tauES_1prong0pizeroDown",
shift_config={"global": {"tau_ES_shift_DM0": 0.998}},
producers={"global": taus.TauPtCorrection},
ignore_producers={"mt": [pairselection.LVMu1, muons.VetoMuons]},
)
)
In this example, a new SystematicShift object is added to the configuration. The name of the shift is tauES_1prong0pizeroDown. The shift_config contains a dictionary of configuration parameters, that are varied for this shift. In this case, the tau_ES_shift_DM0 parameter is set to 0.998. The producers is a dictionary that contains the producers that should be shifted. The ignore_producers is a dictionary that contains the producers that should not be shifted.
In the output file, for all quantities, that depend on the output of the TauPtCorrection producer a shifted version of that quantity will exist as well. The shifted version will be named quantityname__tauES_1prong0pizeroDown, so the name of the shift will be added to the end of the quantity name, separated by __.
Internally, the CROWN framework automatically tracks all shifts and all producers that depend on shifted quantities, and that are not in the ignore_producers list. The shifted quantities are automatically added to the processing and the output file.
Some systematic shifts are already available in the input ROOT file. To incorporate such a shift, a SystematicShiftByQuantity object can be created. The basic structure is
SystematicShiftByQuantity(
name="metUnclusteredEnUp",
quantity_change={
nanoAOD.MET_pt: "PuppiMET_ptUnclusteredUp",
nanoAOD.MET_phi: "PuppiMET_phiUnclusteredUp",
},
scopes=["et", "mt", "tt", "em", "ee", "mm"],
)
In this example, the systematic shift is called metUnclusteredEnUp. The quantity_change is a dictionary that contains the name of the quantity that should be changed and the name of the quantity that should be used as the new quantity. The scopes is a list of the producers that should be affected by this shift.
Modification Rules
Modification rules are used to modify the set of producers that are run. A typical use case for this is, that MC samples require some correction, that does not have to be applied for data. In CROWN, the user would add the corresponding producer to the set of producers to be run, and add a modification rule to the configuration, which removes the corresponding producer for data. A ProducerRule is added using add_modification_rule(). Currently, two types of modification rules exist:
RemoveProducer- Removes a producer from the set of producers to be run.AppendProducer- Appends a producer to the set of producers to be run.
The basic structure is
configuration.add_modification_rule(
["mt", "mm"],
RemoveProducer(producers=scalefactors.MuonIDIso_SF, samples="data"),
)
In this example, the producer MuonIDIso_SF is removed from the set of producers to be run for the sample data. This is done in the mt and mm scope.